6.16. Delta Lake#
6.16.1. Version Your Pandas DataFrame with Delta Lake#
Show code cell content
!pip install deltalake
Versioning your data is essential to undoing mistakes, preventing data loss, and ensuring reproducibility. Delta Lake makes it easy to version pandas DataFrames and review past changes for auditing and debugging purposes.
To version a pandas DataFrame with Delta Lake, start with writing out a pandas DataFrame to a Delta table.
import pandas as pd
import os
from deltalake.writer import write_deltalake
df = pd.DataFrame({"x": [1, 2, 3]})
# Write to a delta table
table = "delta_lake"
os.makedirs(table, exist_ok=True)
write_deltalake(table, df)
Delta Lake stores the data in a Parquet file and maintains a transaction log that records the data operations, enabling time travel and versioning.
delta_lake:
├── 0-4719861e-1d3a-49f8-8870-225e4e46e3a0-0.parquet
└── _delta_log/
│ └──── 00000000000000000000.json
To load the Delta table as a pandas DataFrame, simply use the DeltaTable object:
from deltalake import DeltaTable
dt = DeltaTable(table)
dt.to_pandas()
| x | |
|---|---|
| 0 | 1 |
| 1 | 2 |
| 2 | 3 |
Let’s see what happens when we append another pandas DataFrame to the Delta table.
df2 = pd.DataFrame({"x": [8, 9, 10]})
write_deltalake(table, df2, mode="append")
# Create delta table
dt = DeltaTable(table)
dt.to_pandas()
| x | |
|---|---|
| 0 | 1 |
| 1 | 2 |
| 2 | 3 |
| 3 | 8 |
| 4 | 9 |
| 5 | 10 |
Our Delta table now has two versions. Version 0 contains the initial data and Version 1 includes the data that was appended.
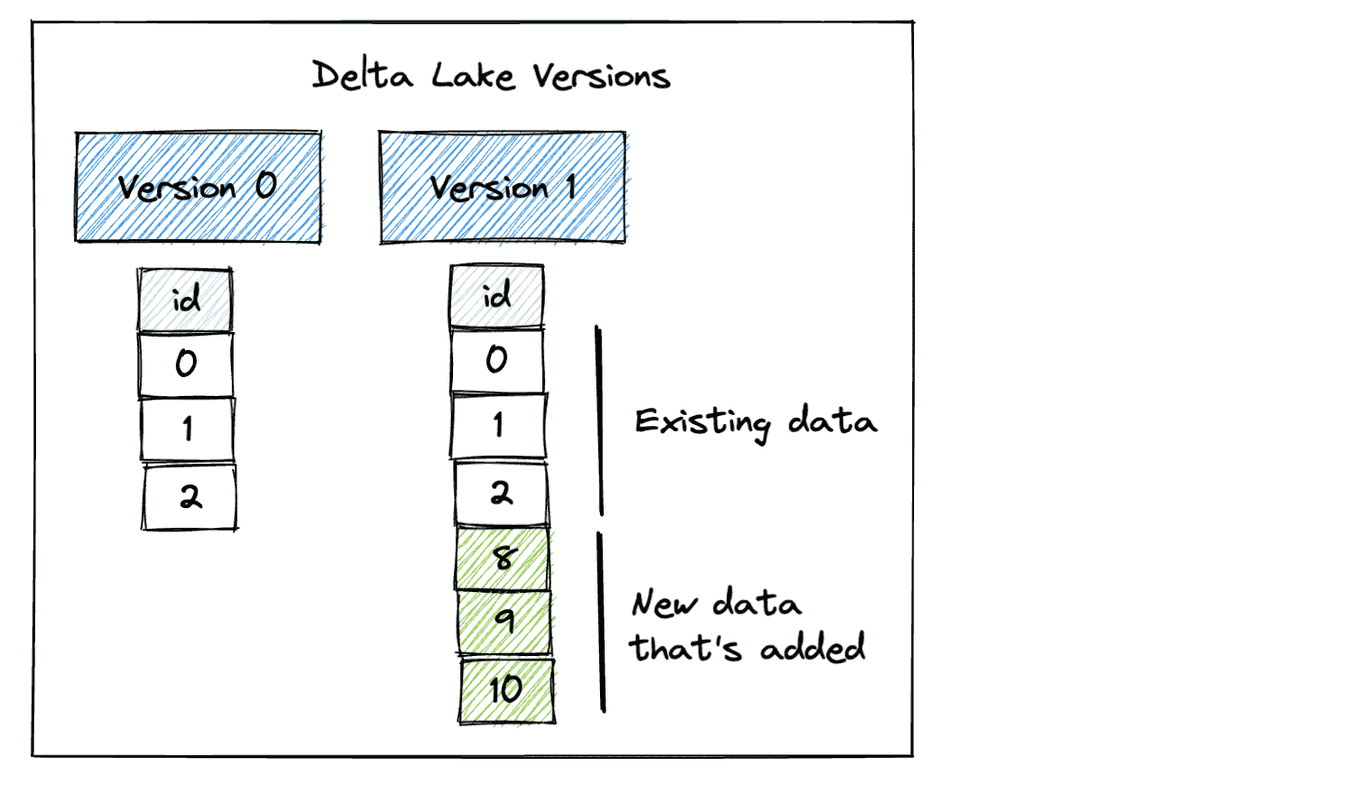
To get the metadata of files that currently make up the current table such as creation time, size, and statistics, call the get_add_actions method.
dt.get_add_actions(flatten=True).to_pandas()
| path | size_bytes | modification_time | data_change | num_records | null_count.x | min.x | max.x | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0-67d190a5-29ed-4555-b946-319769c2226c-0.parquet | 580 | 2024-09-27 21:11:52.351 | True | 3 | 0 | 1 | 3 |
| 1 | 1-24a5cf2f-d7b8-4e0f-8bf5-a12f4d7e2f35-0.parquet | 580 | 2024-09-27 21:11:56.494 | True | 3 | 0 | 8 | 10 |
To access prior versions, simply specify the version number when loading the Delta table:
# Read Version 0 of the dataset
dt0 = DeltaTable(table, version=0)
dt0.to_pandas()
| x | |
|---|---|
| 0 | 1 |
| 1 | 2 |
| 2 | 3 |
6.16.2. Beyond Parquet: Reliable Data Storage with Delta Lake#
Traditional data storage methods, such as plain Parquet files, are susceptible to partial failures during write operations. This can result in incomplete data files and a lack of clear recovery options in the event of a system crash.
Delta Lake’s write operation with ACID transactions helps solve this by:
Ensuring either all data is written successfully or none of it is
Maintaining a transaction log that tracks all changes
Providing time travel capabilities to recover from failures
Here’s an example showing Delta Lake’s reliable write operation:
from deltalake import write_deltalake, DeltaTable
import pandas as pd
initial_data = pd.DataFrame({
"id": [1, 2],
"value": ["a", "b"]
})
write_deltalake("customers", initial_data)
If the append operation fails halfway, Delta Lake’s transaction log ensures that the table remains in its last valid state.
try:
# Simulate a large append that fails halfway
new_data = pd.DataFrame({
"id": range(3, 1003), # 1000 new rows
"value": ["error"] * 1000
})
# Simulate system crash during append
raise Exception("System crash during append!")
write_deltalake("customers", new_data, mode="append")
except Exception as e:
print(f"Write failed: {e}")
# Check table state - still contains only initial data
dt = DeltaTable("customers")
print("\nTable state after failed append:")
print(dt.to_pandas())
# Verify version history
print(f"\nCurrent version: {dt.version()}")
Write failed: System crash during append!
Table state after failed append:
id value
0 1 a
1 2 b
Current version: 0
6.16.3. Optimize Query Speed with Data Partitioning#
Partitioning data allows queries to target specific segments rather than scanning the entire table, which speeds up data retrieval.
The following code uses Delta Lake to select partitions from a pandas DataFrame. Partitioned data loading is approximately 24.5 times faster than loading the complete dataset and then querying a particular subset
import pandas as pd
from deltalake.writer import write_deltalake
from deltalake import DeltaTable
from datetime import datetime
import numpy as np
# Create a DataFrame with hourly sales data for 2 million records
np.random.seed(0) # For reproducibility
start_date = datetime(2023, 1, 1)
end_date = datetime(2023, 8, 31)
date_range = pd.date_range(start_date, end_date, freq='H')
data = {
'datetime': date_range,
'value': np.random.randint(100, 1000, len(date_range))
}
df = pd.DataFrame(data)
df['month'] = df['datetime'].dt.month
df['day'] = df['datetime'].dt.day
df['hour'] = df['datetime'].dt.hour
df[["month", "day", "hour", "value"]].head(5)
| month | day | hour | value | |
|---|---|---|---|---|
| 0 | 1 | 1 | 0 | 784 |
| 1 | 1 | 1 | 1 | 659 |
| 2 | 1 | 1 | 2 | 729 |
| 3 | 1 | 1 | 3 | 292 |
| 4 | 1 | 1 | 4 | 935 |
## Write to a Delta table
table_path = 'delta_lake'
write_deltalake(table_path, df)
%%timeit
## Load the data from the Delta table
DeltaTable(table_path).to_pandas().query("month == 1 & day == 1")
79.2 ms ± 2.62 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
## Write to a Delta table
table_path = "delta_lake2"
write_deltalake(table_path, df, partition_by=["month", "day"])
%%timeit
## Load the data from the Delta table
DeltaTable(table_path).to_pandas([("month", "=", "1"), ("day", "=", "1")])
3.23 ms ± 181 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
6.16.4. Overwrite Partitions of a pandas DataFrame#
Show code cell content
!pip install deltalake
If you need to modify a specific subset of your pandas DataFrame, such as yesterday’s data, it is not possible to overwrite only that partition. Instead, you have to load the entire DataFrame into memory as a workaround solution.
Delta Lake makes it easy to overwrite partitions of a pandas DataFrame.
First, write out a pandas DataFrame as a Delta table that is partitioned by the date column.
import pandas as pd
from deltalake.writer import write_deltalake
from deltalake import DeltaTable
table_path = "tmp/records"
df = pd.DataFrame(
{"a": [1, 2, 3], "date": ["04-21", "04-22", "04-22"]}
)
write_deltalake(
table_path,
df,
partition_by=["date"],
)
The Delta table’s contents are partitioned by date, with each partition represented by a directory
└── _delta_log/
│ └──── 00000000000000000000.json
└── date=04-21/
│ └──── 0-a6813d0c-157b-4ca6-8b3c-8d5afd51947c-0.parquet
└── date=04-22/
│ └──── 0-a6813d0c-157b-4ca6-8b3c-8d5afd51947c-0.parquet
View the Delta table as a pandas DataFrame:
DeltaTable(table_path).to_pandas()
| a | date | |
|---|---|---|
| 0 | 2 | 04-22 |
| 1 | 3 | 04-22 |
| 2 | 1 | 04-21 |
Next, create another DataFrame with two other records on 04-22. Overwrite the 04-22 partition with the new DataFrame and leave other partitions untouched.
df = pd.DataFrame(
{"a": [7, 8], "date": ["04-22", "04-22"]}
)
write_deltalake(
table_path,
df,
mode="overwrite",
partition_filters=[("date", "=", "04-22")],
)
DeltaTable(table_path).to_pandas()
| a | date | |
|---|---|---|
| 0 | 1 | 04-21 |
| 1 | 7 | 04-22 |
| 2 | 8 | 04-22 |
Here is the updated contents of the Delta table:
└── _delta_log/
│ └──── 00000000000000000000.json
│ └──── 00000000000000000001.json
└── date=04-21/
│ └──── 0-a6813d0c-157b-4ca6-8b3c-8d5afd51947c-0.parquet
└── date=04-22/
│ ├──── 0-a6813d0c-157b-4ca6-8b3c-8d5afd51947c-0.parquet
│ └──── 1-b5c9640f-f386-4754-b28f-90e361ab4320-0.parquet
Since the data files are not physically removed from disk, you can time travel to the initial version of the data.
DeltaTable(table_path, version=0).to_pandas()
| a | date | |
|---|---|---|
| 0 | 2 | 04-22 |
| 1 | 3 | 04-22 |
| 2 | 1 | 04-21 |
6.16.5. Efficient Data Appending in Parquet Files: Delta Lake vs. Pandas#
Show code cell content
!pip install deltalake
Appending data to an existing Parquet file using pandas involves:
Loading the entire existing table into memory.
Merging the new data with the existing table.
Writing the merged data to the existing file.
This process can be time-consuming and memory-intensive.
import pandas as pd
df1 = pd.DataFrame([
(1, "John", 5000),
(2, "Jane", 6000),
], columns=["employee_id", "employee_name", "salary"])
df2 = pd.DataFrame([
(3, "Alex", 8000),
], columns=["employee_id", "employee_name", "salary"])
# Save to a parquet file
df1.to_parquet("data.parquet")
## Read the data
existing_data = pd.read_parquet("data.parquet")
## Concat two dataframes
df3 = pd.concat([df1, df2])
# Save to a file
df3.to_parquet("data.parquet")
Delta Lake offers a more efficient approach to handling this process. With Delta Lake, you can add, remove, or modify columns without the need to recreate the entire table.
Delta Lake is also built on top of the Parquet file format so it retains the efficiency and columnar storage benefits of Parquet.
from deltalake.writer import write_deltalake
table_path = "employees"
## Write to Delta Lake
write_deltalake(table_path, df1)
## Append to Delta Lake
write_deltalake(table_path, df2, mode="append")
6.16.6. Safe Data Deletion with delta-rs#
Show code cell content
!pip install deltalake
Data quality management and cleanup operations are critical in modern data lakes. Traditional approaches to data deletion can be problematic:
import pandas as pd
import numpy as np
# Create a large example dataset (1 million rows)
df = pd.DataFrame({
'customer_id': range(1_000_000),
'status': ['active', 'inactive'] * (1_000_000 // 2),
'balance': np.random.uniform(0, 10000, 1_000_000),
})
df.to_parquet('customer_data.parquet')
## Traditional problematic approach
try:
# 1. Load entire file (1M rows) into memory
data = pd.read_parquet('customer_data.parquet') # Memory intensive
# 2. Filter out inactive basic accounts with low balance
data = data[~((data['status'] == 'inactive') &
(data['balance'] < 100))]
# 3. Overwrite file - risk of corruption if process fails
data.to_parquet('customer_data.parquet') # Unsafe
except MemoryError:
print("Not enough memory to process 1M rows")
delta-rs provides a safer approach with transactional guarantees. Here’s how to perform safe deletions:
from deltalake import write_deltalake, DeltaTable
## Write to Delta format
write_deltalake("./customer_data", df)
## Perform safe deletion
dt = DeltaTable("./customer_data")
dt.delete("""
status = 'inactive' AND
balance < 100.0
""")
{'num_added_files': 1,
'num_removed_files': 1,
'num_deleted_rows': 5019,
'num_copied_rows': 994981,
'execution_time_ms': 220121,
'scan_time_ms': 69760,
'rewrite_time_ms': 148}
You can verify the operation and access previous versions:
## Check operation history
print("Operation History:")
for entry in dt.history():
print(f"Version {entry['version']}: {entry['operation']}")
# Access the previous version if needed
dt_previous = DeltaTable("./customer_data", version=dt.version() - 1)
print("\nPrevious version data:")
print(dt_previous.to_pandas().head(10))
Operation History:
Version 1: DELETE
Version 0: CREATE TABLE
Previous version data:
customer_id status balance
0 0 active 2281.154365
1 1 inactive 8330.057614
2 2 active 9908.828674
3 3 inactive 3766.795979
4 4 active 8536.698571
5 5 inactive 8681.325433
6 6 active 1610.351636
7 7 inactive 2537.934431
8 8 active 4278.094055
9 9 inactive 3531.708249
6.16.7. Enforce Data Quality with Delta Lake Constraints#
Delta Lake provides a convenient way to enforce data quality by adding constraints to a table, ensuring that only valid and consistent data can be added.
In the provided code, attempting to add new data with a negative salary violates the constraint of a positive salary, and thus, the data is not added to the table.
Show code cell content
!pip install deltalake
import pandas as pd
from deltalake.writer import write_deltalake
from deltalake import DeltaTable
table_path = "delta_lake"
df1 = pd.DataFrame(
[
(1, "John", 5000),
(2, "Jane", 6000),
],
columns=["employee_id", "employee_name", "salary"],
)
write_deltalake(table_path, df1)
df1
| employee_id | employee_name | salary | |
|---|---|---|---|
| 0 | 1 | John | 5000 |
| 1 | 2 | Jane | 6000 |
table = DeltaTable(table_path)
table.alter.add_constraint({"salary_gt_0": "salary > 0"})
df2 = pd.DataFrame(
[(3, "Alex", -200)],
columns=["employee_id", "employee_name", "salary"],
)
write_deltalake(table, df2, mode="append", engine="rust")
DeltaProtocolError: Invariant violations: ["Check or Invariant (salary > 0) violated by value in row: [3, Alex, -200]"]
6.16.8. Efficient Data Updates and Scanning with Delta Lake#
Show code cell content
!pip install -U "deltalake==0.10.1"
Every time new data is appended to an existing Delta table, a new Parquet file is generated. This allows data to be ingested incrementally without having to rewrite the entire dataset.
As files accumulate, read operations may surge. The compact function merges small files into larger ones, enhancing scanning performance.
Combining incremental processing with the compact function enables efficient data updates and scans as your dataset expands.
import pandas as pd
from deltalake.writer import write_deltalake
table_path = 'delta_lake'
data_url = "https://gist.githubusercontent.com/khuyentran1401/458905fc5c630d7a1f7a510a04e5e0f9/raw/5b2d760011c9255a68eb08b83b3b8759ffa25d5c/data.csv"
dfs = pd.read_csv(data_url, chunksize=100)
for df in dfs:
write_deltalake(table_path, df, mode="append")
from deltalake import DeltaTable
dt = DeltaTable(table_path)
%%timeit
df = dt.to_pandas()
30.6 ms ± 2.94 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
dt.optimize.compact()
{'numFilesAdded': 1,
'numFilesRemoved': 100,
'filesAdded': {'min': 278115,
'max': 278115,
'avg': 278115.0,
'totalFiles': 1,
'totalSize': 278115},
'filesRemoved': {'min': 5712,
'max': 5717,
'avg': 5715.8,
'totalFiles': 100,
'totalSize': 571580},
'partitionsOptimized': 1,
'numBatches': 100,
'totalConsideredFiles': 100,
'totalFilesSkipped': 0,
'preserveInsertionOrder': True}
%%timeit
df = dt.to_pandas()
1.32 ms ± 49 µs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
6.16.9. Simplify Table Merge Operations with Delta Lake#
Show code cell content
!pip install delta-spark
Merging two datasets and performing both insert and update operations can be a complex task.
Delta Lake makes it easy to perform multiple data manipulation operations during a merge operation.
The following code demonstrates merging two datasets using Delta Lake:
If a match is found, the
last_talkcolumn inpeople_tableis updated with the corresponding value fromnew_df.If the
last_talkvalue inpeople_tableis older than 30 days and the corresponding row is not present in thenew_dftable, thestatuscolumn is updated to ‘rejected’.
Show code cell content
import pyspark
from delta import *
## Configure Spark to use Delta
builder = (
pyspark.sql.SparkSession.builder.appName("MyApp")
.config("spark.sql.extensions", "io.delta.sql.DeltaSparkSessionExtension")
.config(
"spark.sql.catalog.spark_catalog",
"org.apache.spark.sql.delta.catalog.DeltaCatalog",
)
)
spark = configure_spark_with_delta_pip(builder).getOrCreate()
:: loading settings :: url = jar:file:/Users/khuyentran/book/venv/lib/python3.11/site-packages/pyspark/jars/ivy-2.5.1.jar!/org/apache/ivy/core/settings/ivysettings.xml
Ivy Default Cache set to: /Users/khuyentran/.ivy2/cache
The jars for the packages stored in: /Users/khuyentran/.ivy2/jars
io.delta#delta-spark_2.12 added as a dependency
:: resolving dependencies :: org.apache.spark#spark-submit-parent-3f072ef1-cd28-41e1-8ccd-8da112101571;1.0
confs: [default]
found io.delta#delta-spark_2.12;3.2.0 in central
found io.delta#delta-storage;3.2.0 in central
found org.antlr#antlr4-runtime;4.9.3 in central
:: resolution report :: resolve 295ms :: artifacts dl 18ms
:: modules in use:
io.delta#delta-spark_2.12;3.2.0 from central in [default]
io.delta#delta-storage;3.2.0 from central in [default]
org.antlr#antlr4-runtime;4.9.3 from central in [default]
---------------------------------------------------------------------
| | modules || artifacts |
| conf | number| search|dwnlded|evicted|| number|dwnlded|
---------------------------------------------------------------------
| default | 3 | 0 | 0 | 0 || 3 | 0 |
---------------------------------------------------------------------
:: retrieving :: org.apache.spark#spark-submit-parent-3f072ef1-cd28-41e1-8ccd-8da112101571
confs: [default]
0 artifacts copied, 3 already retrieved (0kB/12ms)
24/09/29 14:32:49 WARN NativeCodeLoader: Unable to load native-hadoop library for your platform... using builtin-java classes where applicable
Setting default log level to "WARN".
To adjust logging level use sc.setLogLevel(newLevel). For SparkR, use setLogLevel(newLevel).
Show code cell content
# Create a spark dataframe
data = [
(0, "A", "2023-04-15", "interviewing"),
(1, "B", "2023-05-01", "interviewing"),
(2, "C", "2023-03-01", "interviewing"),
]
df = (
spark.createDataFrame(data)
.toDF("id", "company", "last_talk", "status")
.repartition(1)
)
## Write to a delta table
path = "tmp/interviews"
df.write.format("delta").save(path)
Show code cell content
from delta.tables import DeltaTable
## Update the delta table
people_table = DeltaTable.forPath(spark, path)
## Target table
people_table.toDF().show()
24/09/29 14:33:18 WARN SparkStringUtils: Truncated the string representation of a plan since it was too large. This behavior can be adjusted by setting 'spark.sql.debug.maxToStringFields'.
+---+-------+----------+------------+
| id|company| last_talk| status|
+---+-------+----------+------------+
| 0| A|2023-04-15|interviewing|
| 1| B|2023-05-01|interviewing|
| 2| C|2023-03-01|interviewing|
+---+-------+----------+------------+
Show code cell content
new_data = [(0, "A", "2023-05-07")]
new_df = (
spark.createDataFrame(new_data).toDF("id", "company", "last_talk").repartition(1)
)
## Source table
new_df.show()
+---+-------+----------+
| id|company| last_talk|
+---+-------+----------+
| 0| A|2023-05-07|
+---+-------+----------+
one_month_ago = "current_date() - INTERVAL '30' DAY"
people_table.alias("target").merge(
new_df.alias("source"), "target.id = source.id"
).whenMatchedUpdate(
set={"target.last_talk": "source.last_talk", "target.status": "'interviewing'"}
).whenNotMatchedBySourceUpdate(
condition=f"target.last_talk <= {one_month_ago}",
set={"target.status": "'rejected'"},
).execute()
people_table.toDF().show()
+---+-------+----------+------------+
| id|company| last_talk| status|
+---+-------+----------+------------+
| 0| A|2023-05-07|interviewing|
| 1| B|2023-05-01|interviewing|
| 2| C|2023-03-01| rejected|
+---+-------+----------+------------+
6.16.10. From Complex SQL to Simple Merges: Delta Lake’s Upsert Solution#
Show code cell content
!pip install delta-spark
Traditionally, implementing upsert (update or insert) logic requires separate UPDATE and INSERT statements or complex SQL. This approach can be error-prone and inefficient, especially for large datasets.
Delta Lake’s merge operation solves this problem by allowing you to specify different actions for matching and non-matching records in a single, declarative statement.
Here’s an example that demonstrates the power and simplicity of Delta Lake’s merge operation:
First, let’s set up our initial data:
# Create sample data for 'customers' DataFrame
customers_data = [
(1, "John Doe", "john@example.com", "2023-01-01 10:00:00"),
(2, "Jane Smith", "jane@example.com", "2023-01-02 11:00:00"),
(3, "Bob Johnson", "bob@example.com", "2023-01-03 12:00:00"),
]
customers = spark.createDataFrame(
customers_data, ["customer_id", "name", "email", "last_updated"]
)
# Create sample data for 'updates' DataFrame
updates_data = [
(2, "Jane Doe", "jane.doe@example.com"), # Existing customer with updates
(3, "Bob Johnson", "bob@example.com"), # Existing customer without changes
(4, "Alice Brown", "alice@example.com"), # New customer
]
updates = spark.createDataFrame(updates_data, ["customer_id", "name", "email"])
## Show the initial data
print("Initial Customers:")
customers.show()
print("Updates:")
updates.show()
Initial Customers:
+-----------+-----------+----------------+-------------------+
|customer_id| name| email| last_updated|
+-----------+-----------+----------------+-------------------+
| 1| John Doe|john@example.com|2023-01-01 10:00:00|
| 2| Jane Smith|jane@example.com|2023-01-02 11:00:00|
| 3|Bob Johnson| bob@example.com|2023-01-03 12:00:00|
+-----------+-----------+----------------+-------------------+
Updates:
+-----------+-----------+--------------------+
|customer_id| name| email|
+-----------+-----------+--------------------+
| 2| Jane Doe|jane.doe@example.com|
| 3|Bob Johnson| bob@example.com|
| 4|Alice Brown| alice@example.com|
+-----------+-----------+--------------------+
Next, we create a Delta table from our initial customer data:
## Define the path where you want to save the Delta table
delta_table_path = "customers_delta"
## Write the DataFrame as a Delta table
customers.write.format("delta").mode("overwrite").save(delta_table_path)
# Create a DeltaTable object
customers_delta = DeltaTable.forPath(spark, delta_table_path)
print("Customers Delta Table created successfully")
Customers Delta Table created successfully
Now, here’s the key part - the merge operation that handles both updates and inserts in a single statement:
## Assume 'customers_delta' is your target table and 'updates' is your source of new data
customers_delta.alias("target").merge(
updates.alias("source"),
"target.customer_id = source.customer_id"
).whenMatchedUpdate(set={
"name": "source.name",
"email": "source.email",
"last_updated": "current_timestamp()"
}).whenNotMatchedInsert(values={
"customer_id": "source.customer_id",
"name": "source.name",
"email": "source.email",
"last_updated": "current_timestamp()"
}).execute()
## Verify the updated data
print("Updated Customers Delta Table:")
customers_delta.toDF().show()
Updated Customers Delta Table:
+-----------+-----------+--------------------+--------------------+
|customer_id| name| email| last_updated|
+-----------+-----------+--------------------+--------------------+
| 2| Jane Doe|jane.doe@example.com|2024-09-29 14:34:...|
| 3|Bob Johnson| bob@example.com|2024-09-29 14:34:...|
| 4|Alice Brown| alice@example.com|2024-09-29 14:34:...|
| 1| John Doe| john@example.com| 2023-01-01 10:00:00|
+-----------+-----------+--------------------+--------------------+
6.16.11. The Best Way to Append Mismatched Data to Parquet Tables#
Appending mismatched data to a Parquet table involves reading the existing data, concatenating it with the new data, and overwriting the existing Parquet file. This approach can be expensive and may lead to schema inconsistencies.
In the following code, the datatype of col3 is supposed to be int64 instead of float64.
import pandas as pd
filepath = 'test.parquet'
## Write a dataframe to a parquet file
df1 = pd.DataFrame({'col1': [1, 2], 'col2': [3, 4]})
df1.to_parquet(filepath)
## Append a dataframe to a parquet file
df2 = pd.DataFrame({'col1': [2], 'col2': [7], 'col3': [0]})
concatenation = pd.concat([df1, df2]) # concatenate dataframes
concatenation.to_parquet(filepath) # overwrite original file
concat_df = pd.read_parquet(filepath)
print(concat_df, "\n")
print(concat_df.dtypes)
col1 col2 col3
0 1 3 NaN
1 2 4 NaN
0 2 7 0.0
col1 int64
col2 int64
col3 float64
dtype: object
With Delta Lake, you can effortlessly append DataFrames with extra columns while ensuring the preservation of your data’s schema.
Show code cell content
import pyspark
from delta import *
## Configure Spark to use Delta
builder = (
pyspark.sql.SparkSession.builder.appName("MyApp")
.config("spark.sql.extensions", "io.delta.sql.DeltaSparkSessionExtension")
.config(
"spark.sql.catalog.spark_catalog",
"org.apache.spark.sql.delta.catalog.DeltaCatalog",
)
)
spark = configure_spark_with_delta_pip(builder).getOrCreate()
# Create a spark Dataframe
data = [(1, 3), (2, 4)]
df1 = (
spark.createDataFrame(data)
.toDF("col1", "col2")
.repartition(1)
)
## Write to a delta table
path = "tmp"
df1.write.format("delta").save(path)
# Create a new DataFrame
new_data = [(2, 7, 0)]
df2 = (
spark.createDataFrame(new_data).toDF("col1", "col2", "col3").repartition(1)
)
df2.show()
+----+----+----+
|col1|col2|col3|
+----+----+----+
| 2| 7| 0|
+----+----+----+
## Append to the existing Delta table
df2.write.option("mergeSchema", "true").mode("append").format("delta").save(path)
## Read the Delta table
from delta.tables import DeltaTable
table = DeltaTable.forPath(spark, path)
concat_df = table.toDF().pandas_api()
print(concat_df, "\n")
print(concat_df.dtypes)
col1 col2 col3
0 2 7 0.0
1 1 3 NaN
2 2 4 NaN
col1 int64
col2 int64
col3 int64
dtype: object
